20.05.2013
For the upcoming Bio7 1.7 release i worked on a new general shell implementation. And with this implementation some new ideas in conjunction with R came to my mind.
So recently i integrated some new actions to ease the creation of R packages with R.
Please note: For the built process of R package binaries some tools are needed (especially for Windows). Please consult the R instructions on the internet or search for available tutorials.
For tests on the Windows OS i installed a Latex environment (MiKeTeX with inconsolata.sty style file installed) and the Rtools for Windows.
With the upcoming Bio7 release the following actions will be available:
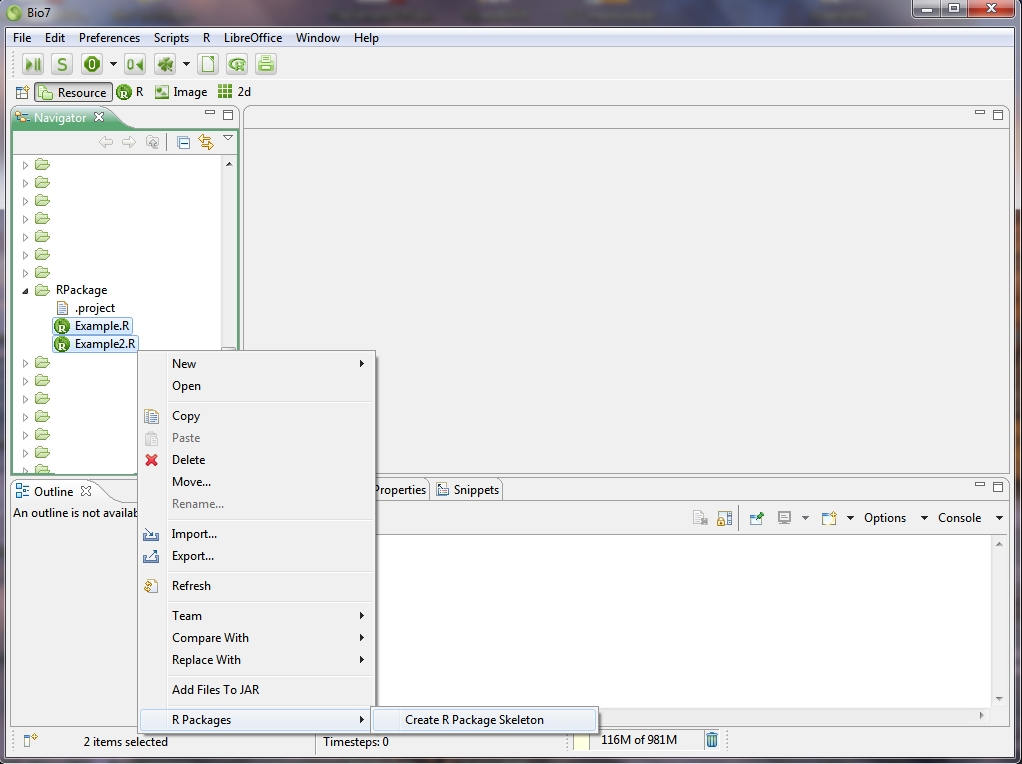
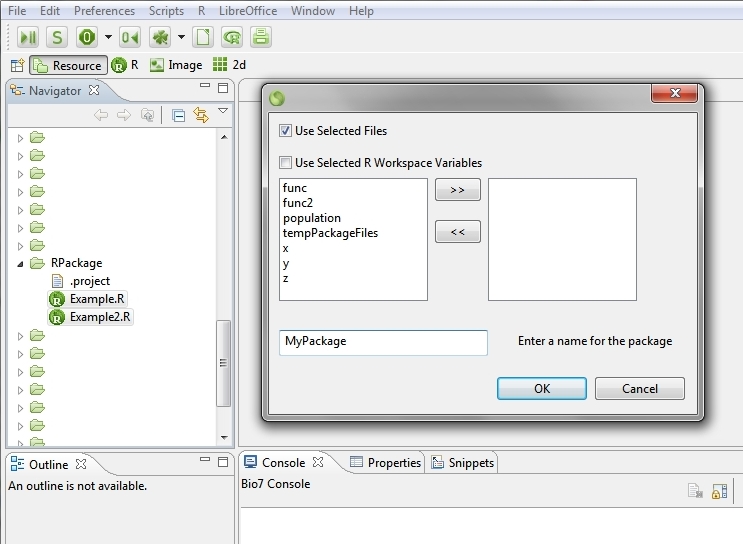
In the file Navigator of Bio7 you can create a package skeleton from select R files or R workspace variables. By invoking the Create R Package Skeleton action a dialog pops up which offers options to create a package skeleton from the selected files or from availabe workspace variables and to enter a name for the package (see the next two screenshots below).
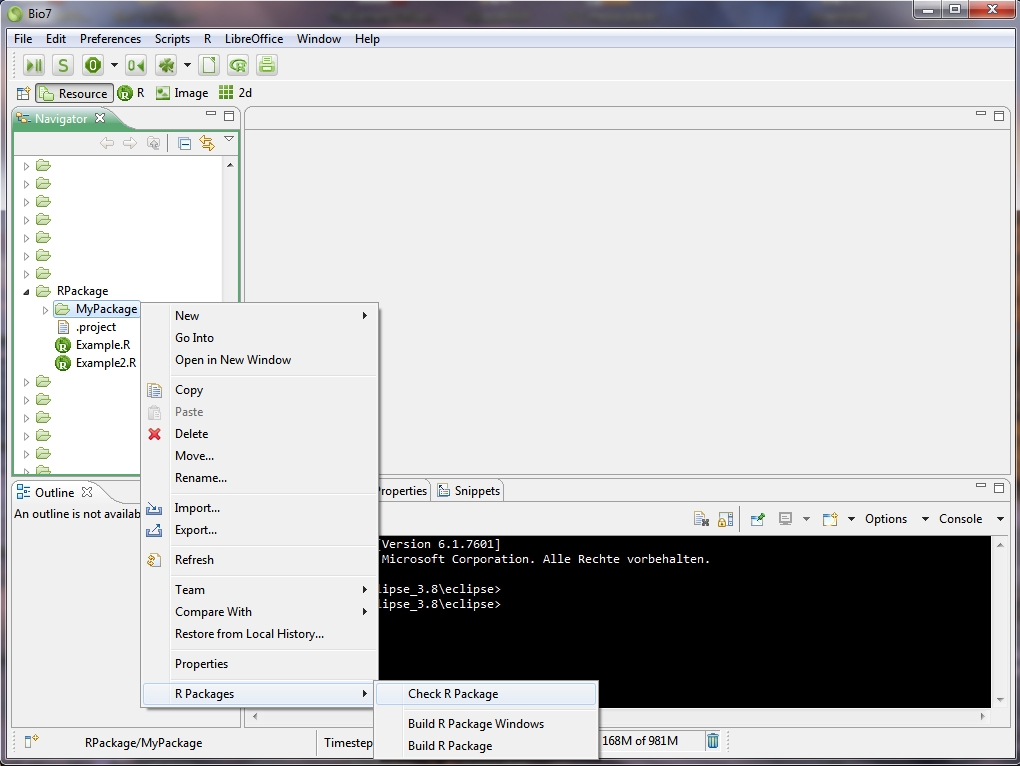
After the confirmation with “OK” the R package skeleton can be found as a subfolder of the selected Bio7 project. If you now select the created R skeleton folder and open the context menu of the file Navigator a new submenu is available with actions to check and built the R package (see screenshot below)
For the built process the new available native shell hast to be selected from the console menu of the Console view of Bio7 1.7.
If everything is configured properly (see the shell messages after the check or built process) a new built R package should be available in the R package folder (after a refresh of the folder).