07.02.2020
A new release of Bio7 is available. This update comes with a plethora of new features, improvements and bugfixes.
For those who don’t know Bio7. The application Bio7 is an integrated development environment for ecological modeling, scientific image analysis and statistical analysis.
It also contains a feature complete development environment for R with an advanced R editor, R developer tools and interfaces to perform scientific image analysis with R and the also embedded ImageJ application.
New and Noteworthy
General:
- Bio7 3.1 RCP (Rich Client Platform) built upon Eclipse 4.14
- Bundled with AdoptOpenJDK 13.0.2
- Bundled with R 3.6.2 (Windows only!)
- New main toolbar action available to create different projects and files in one shot
- Added a toolbar submenu action to open the ‘Control’ view
- ‘Dark Mode’ theme automatically enabled on supported OS (MacOSX only!)
- ‘2D’ perspective (e.g., for cellular automata simulations) is now hidden by default.
- Removed the ‘Image’ perspective and the ‘Points Panel’ view and actions
- Updated libraries (SSH for Windows, etc.)
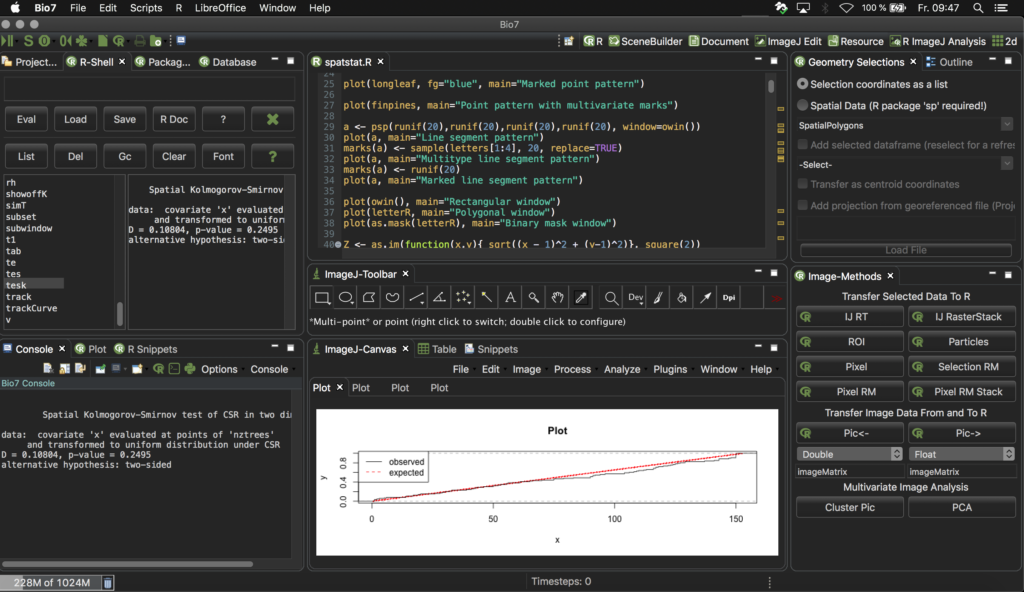
- Improved the ‘WorldWind’, ‘2D’ and ‘3D’ perspective to open and arrange the editor area in a user-friendly programming layout by default (see screenshots below).
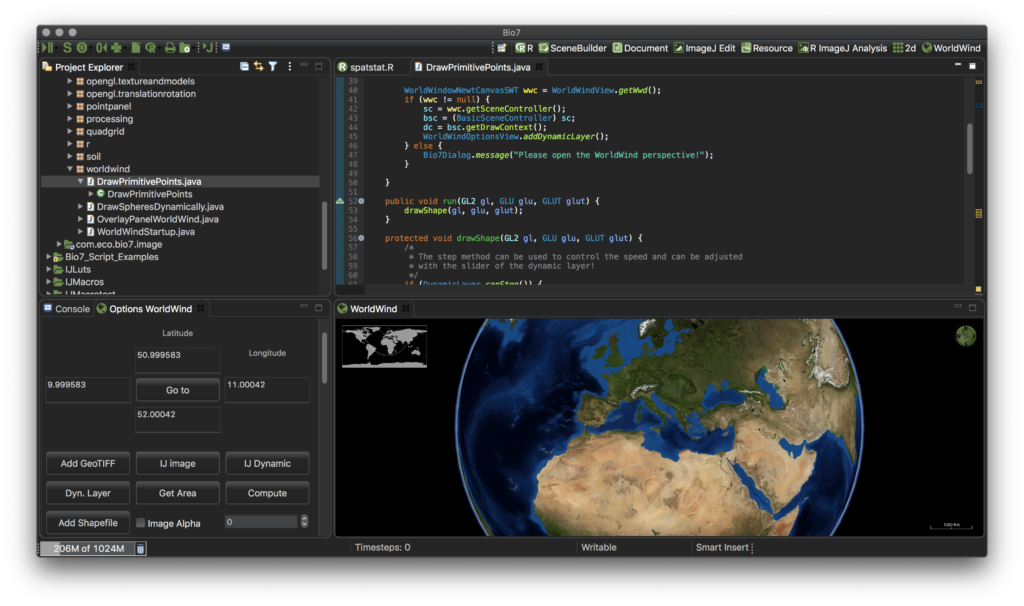
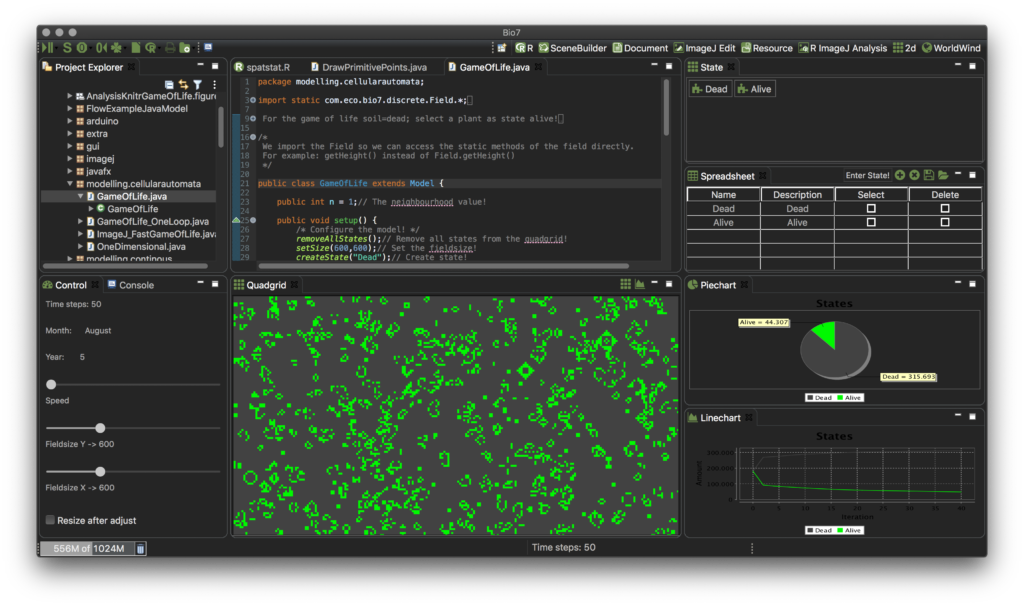
- JavaFX has been disabled by default in the preferences for Linux HighDPI displays (e.g., GTK scale 2) because of an open bug when embedded in an SWT canvas.
- Improved the default Bio7 CSS interface colors
R:
- New toolbar wizard to create quickly R, R Markdown and Shiny files in a project
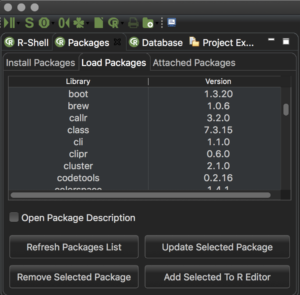
- New R Packages tab available to show and update installed R packages (see screenshot below)
- Changed the ‘Image-Methods’ view to show only R/ImageJ related actions (bidirectional transfer of image data, selection data, particle and cluster methods, etc.)
- Added the ‘Geometry Selections’ view to the ‘R ImageJ Analysis’ perspective to transfer ROI selection coordinates as a list or georeferenced data geometries (using package ‘sp’ and Java GDAL)
- Added the KMeans cluster algorithm to the ‘Cluster Pic’ action in the ‘Image-Methods’ view
- The ‘R ImageJ Analysis’ perspective is now visible by default
- Improved the ‘Pixel RM’ and ‘Pixel RM Stack’ action to transfer all ROI selections more efficiently, especially for VirtualStacks of ImageJ
- Added a new option in the ‘Pixel RM’ and ‘Pixel RM Stack’ action to transfer ROI Manager selections signature names according to a separator char (handy for supervised classification with ImageJ and R)
- Added a new stack transfer API method (‘imageFeatureStackToR’) to transfer whole stacks in a column layout (for pixel classification)
- The ‘R-Shell’ view list now scrolls to the last selected variables after a workspace refresh
- New R editor ‘Quickfixes’ available (warning for an assignment operator in function call, check for null and na in comparison operators)
- Improved the code completion of the ‘R-Shell’ view and the R editor
- Improved the plot layout in ImageJ (when using the preference option ‘ImageJ View Display Size’ in the Rserve plot preferences)
- Added a Shiny timeout to improve the server startup
- Updated the R ANTLR parser
ImageJ:
- Updated ImageJ to version 1.52t(.51)
- Added a new ImageJ macro debugger interface
- Added a new thumbnail action to the context menu of the ‘Navigator’ view to open image and LUT files of a selected directory
- Added a ‘Detach All Images’ action to transfer all opened tab images to a view (which can be moved and arranged like a separate window)
- Improved the dark theme for all AWT components
- Added new default macro editor templates
- Improved the ImageJ plot windows
- Improved the detached image view layout
- Improved the canvas layout for HighDPI displays on Windows
- Huge amount of other changes since the last release. For all changes, see:https://github.com/Bio7/EclipseImageJ1Plugin
Java:
- Dynamic compilation now supports Java 13 by default
- Added a new option to dynamically compile a class without a constructor call (without object creation). The object has to be created in the main method (as known from Java).
- Updated the Java OpenGL (JOGL) 3D interfaces
- Updated Java WorldWind to the latest version
- Fixed the OpenGL view visibility on MacOSX (the ‘3D’ and ‘WorldWind’ view stayed visible when switching to another perspective)
- Added an experimental fullscreen option to open WorldWind and the ‘3D’ view on a secondary monitor (press F3)
- Updated Java libraries
Download and Installation:
Windows:
Just download the *.zip distribution file from https://bio7.org and unzip it in your preferred location. Bio7 comes bundled with a Java Runtime Environment, R and Rserve distribution and works out of the box.
Linux:
Download and extract the installation file from https://bio7.org.
For Linux you have to install R and Rserve (see Rserve installation below!).
MacOSX:
Download and extract the installation file from https://bio7.org.
If you start Bio7 a warning or error can occur because of the changes how Apple treats signatures! To allow Bio7 to start see this instructions for Yosemite, Sierra and Mojave:
First try to open the app with the context menu to allow the execution. If that won’t work try the following:
Yosemite: Open an app from an unidentified developer
Sierra: Open an app from an unidentified developer
In addition for MacOSX you have to install R and Rserve (see below!).
Linux and MacOSX Rserve (compiled for cooperative mode) installation:
To install Rserve open the ‘Native R’ console in the ‘Console’ view and then execute the view menu action “Options -> Install Rserve (coop. mode) for R …” for different R versions (SSL 1.1 version for Linux Ubuntu > 19.10). This will download and install Rserve in your default R library location, see video below (please make sure that your default Linux R library install location has writing permissions!):
How to install Rserve for Linux and MacOSX: https://youtu.be/tF7HbRBRIF
In cooperative mode only one connection at a time is allowed (which we want for this Desktop appl.) and all subsequent connections share the same namespace (default on Windows)!
Bio7 Documentation
For more information about Bio7 please consult the soon updated Bio7 User Guide.
A plethora of Bio7 videotutorials for an introduction can be found on YouTube.