29.08.2018
Recently I discovered the package PyRserve for Python which connects Python with R using Rserve.
This is extremly useful because Bio7 already integrates Rserve and has special GUI interfaces available to transfer, e.g., data from spreadsheets, ImageJ image and selection data (also georeferenced), Java simulation data, etc.
With this new Rserve connection this data can now easily transferred from Java and R to a Python workflow.
Python as a language is already supported in Bio7 with an integration of Jython (which runs on the Java platform a supported JVM language like Groovy, JavaScript, Scala, etc. and has a direct access to the Bio7 API).
In additon native Python can be executed with the default Bio7 Python editor if configured in the native preferences of Bio7 (running as an external process in a Bio7 Python shell if enabled).
However since Bio7 is based on Eclipse a more powerful editor for Python/Jython can be installed with PyDev which is a complete Python IDE for Eclipse and available as an Eclipse plugin which can be installed with the Bio7 (Eclipse) Update Manager
Here a video how PyDev can be installed in Bio7:
Update Site PyDev: http://www.pydev.org/updates/
If PyDev is installed you can create Python projects in Bio7 and execute Python and Jython scripts depending on the settings in it’s own process (e.g. with the action editor context menu-> Run As->Python Run) .
In addition it is possible to use a special Bio7 toolbar action (visible if the PyDev editor or Bio7 Python editor is opened) to interpret the content of the PyDev editor in the current Bio7 Jython or Python process (which is of no importance for the following use of PyRserve!).
PyRserve can be installed easily using the pip command in the python shell:
pip install PyRserve
or by using PyDev with the preferences dialog (see screenshot below).
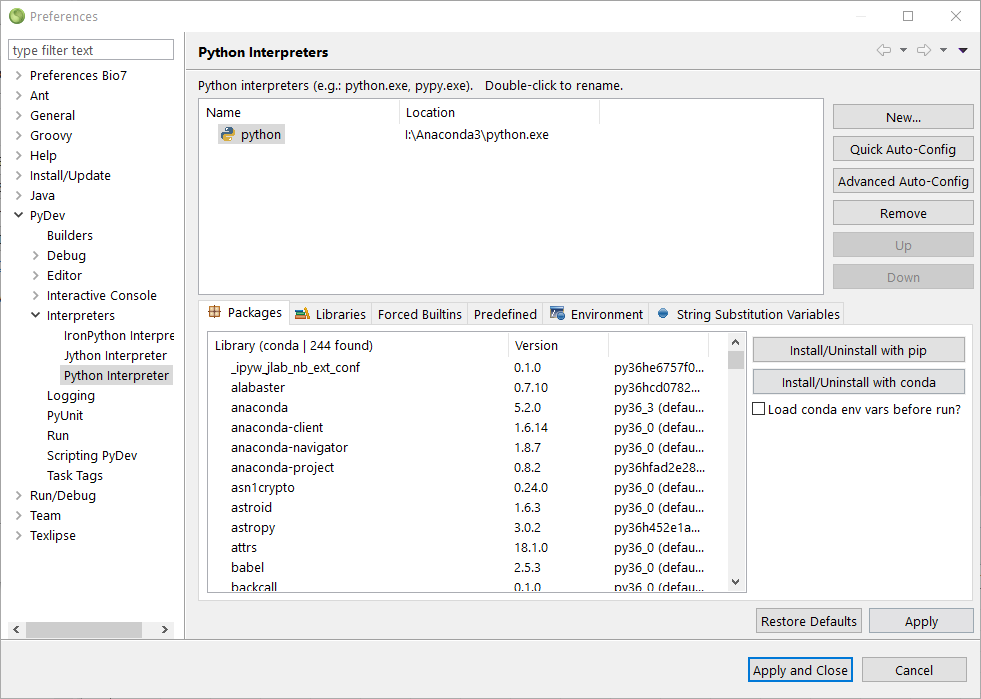
After the installation we can start RServe from within R as usual ,e.g., with:
library(Rserve)
run.Rserve()
To connect Python to Rserve the following commands are suffcient:
import pyRserve
conn = pyRserve.connect()
More examples can be found in the PyRserve documentation.
However since the Bio7 functionality can be easily extended with scripts (which can be added dynamically to a menu copying them to a special scripts location, see screenshot below) I decided to write two short Groovy scripts to start and stop Rserve not controlled by the Bio7 Java connection interface (and if necessary disconnect R from a current Bio7 controlled Java connection).
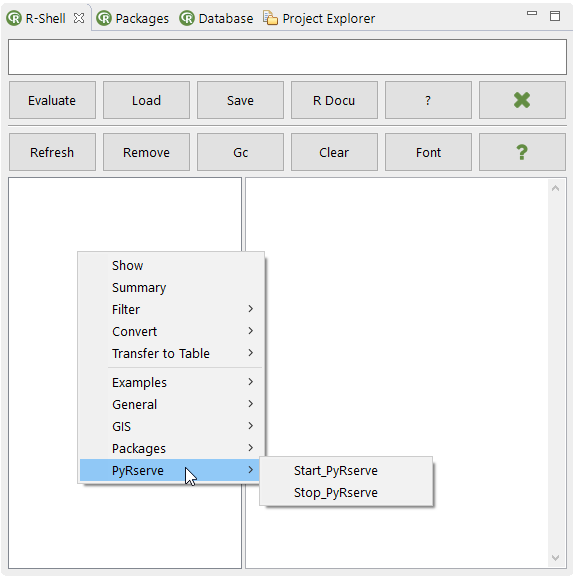
The ‘Start_PyRserve’ script starts R and Rserve if not alive in the Bio7 R console and also disconnects from a running Java Rserve connection if activated.
Note that in Bio7 only one connection to Rserve is allowed (Rserve cooperative mode).
The ‘Stop_PyRserve’ script reconnects Python (closed in the Python scripts!) to shutdown Rserve and returns to the Bio7 console and R (Pydev has it’s own console).
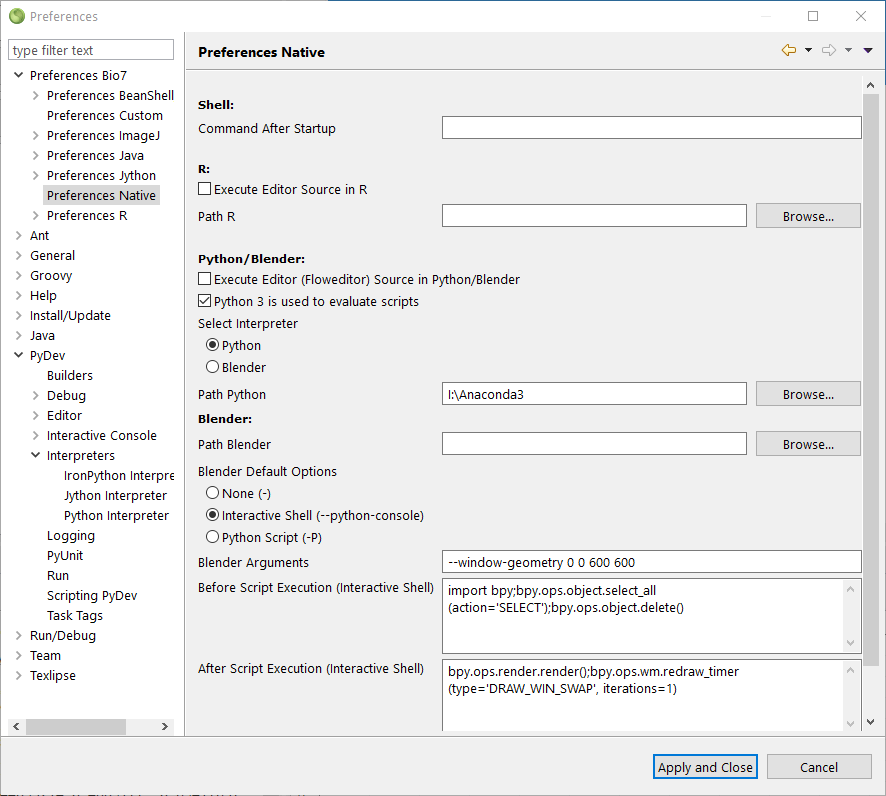
This shutdown command is excecuted by an external Python process. To get the path for the Python interpreter we also have to set the Bio7 Python path (not PyDev!) in the Bio7 preferences (see Windows screenshot below – normally not necessary on Linux or when the Python interpreter is on the OS PATH!).
With this two scripts it is now (more secure) possible to start Rserve and use the Bio7 Java interface, e.g., to transfer image data to R (ImageJ to R), start the Python connection (which disconnects Java) and use the image data inside a Python workflow (see the second example).
Here a first example script to transfer random numbers (in PyDev use the context menu action ‘Runs As->Python Run’ of the editor to execute the Python script) with a PyRserve connection.
import pyRserve
conn = pyRserve.connect()
result = conn.eval("result<-runif(100)")
conn.close()
print (result)
Here another example script to plot a transferred image matrix in Python with matplotlib (8-bit, grayscale as double matrix) if available in the R workspace: (see this old video how to do this with Bio7):
import pyRserve
import matplotlib.pylab as plt
conn = pyRserve.connect()
matrix = conn.eval("tryCatch({matrix<-imageMatrix}, error = function(e) {return ('NULL')})")
conn.close()
if matrix != 'NULL':
# we have to transpose the image for a correct view!
matrix = matrix.T
plt.imshow(matrix)
plt.colorbar()
plt.show()
Here a summary of the installation and configuration steps necessary for Bio7 to use PyRserve and execute Python scripts with the powerful PyDev editor:
- Install the PyDev editor and configure the Python path
- Install PyRserve (pip install PyRserve)
- Configure the Python path of Bio7 (Preferences Bio7->Preferences Native)
- Install the Groovy Start/Stop scripts for PyRserve (simply unzip Examples_PyRserve.zip in one scripts location – Open ‘Scripts->General->Open_Script_Location’ from the main Bio7 menu or in the context menu of the R-Shell view open General->Open_Script_Location’. Put them in an extra folder for a submenu!
- Start the ‘Start_PyRserve’ script to start Rserve (not Bio7 controlled!)
- Execute a Python script which connects and disconnects PyRserve
- Stop the server and shutdown Rserve with the ‘Stop_PyRserve’ script
Download Start/Stop PyRserve scripts here