17.10.2017
A new version of Bio7 for Windows and Linux is available. The MacOSX version will be released soon, too.
The application Bio7 is an integrated development environment for ecological modelling, scientific image analysis and statistical analysis. Beside many different tools it contains a complete Graphical User Interface for R.
The application itself is based on an RCP-Eclipse-Environment (Rich-Client-Platform) which offers a huge flexibility in configuration and extensibility because of its plug-in structure and the possibility of customization.
For a short overview see this older video.
New Features:
General
- Improved the dark theme for Bio7 2.6 (see Windows and Linux screenshot below)
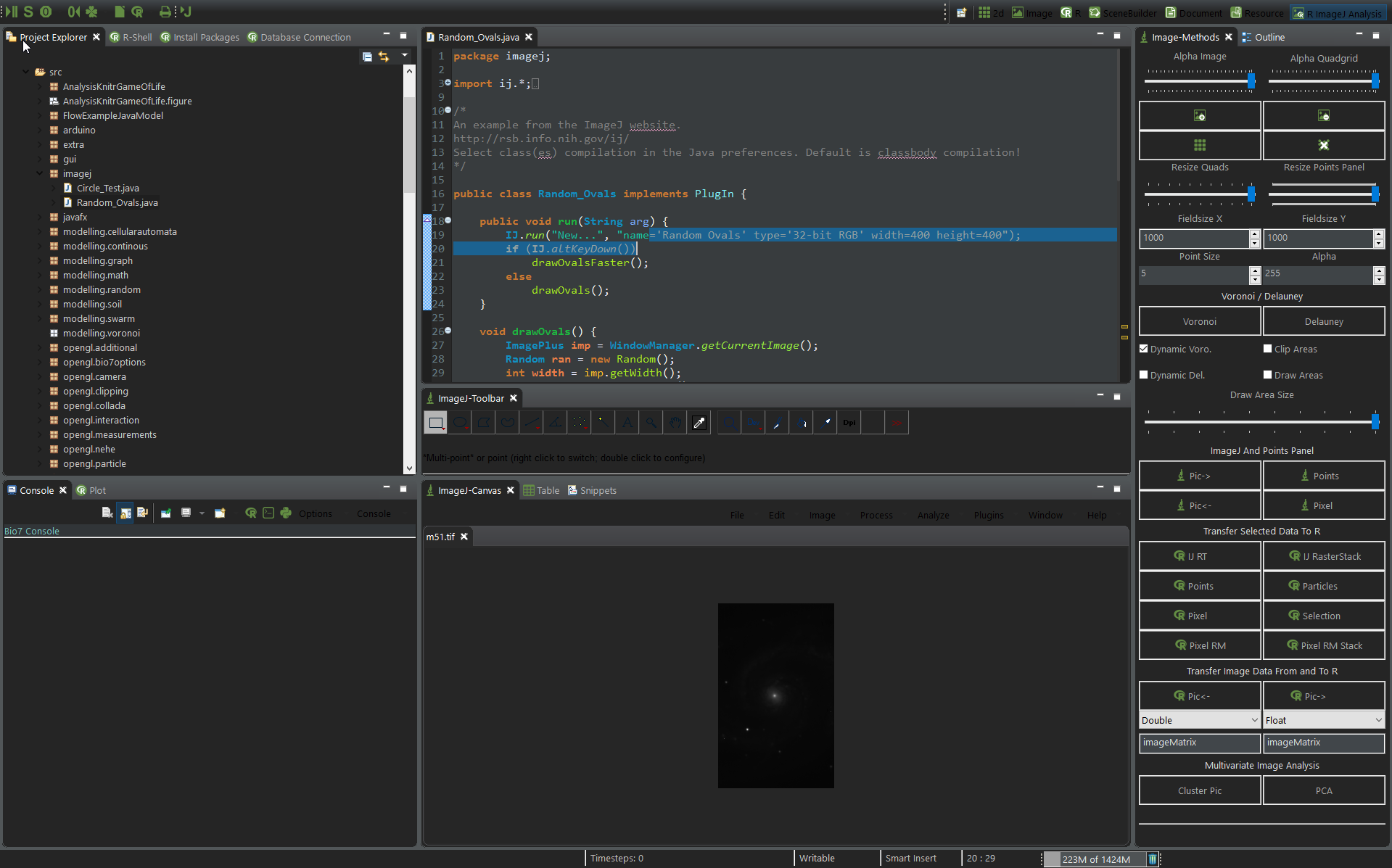
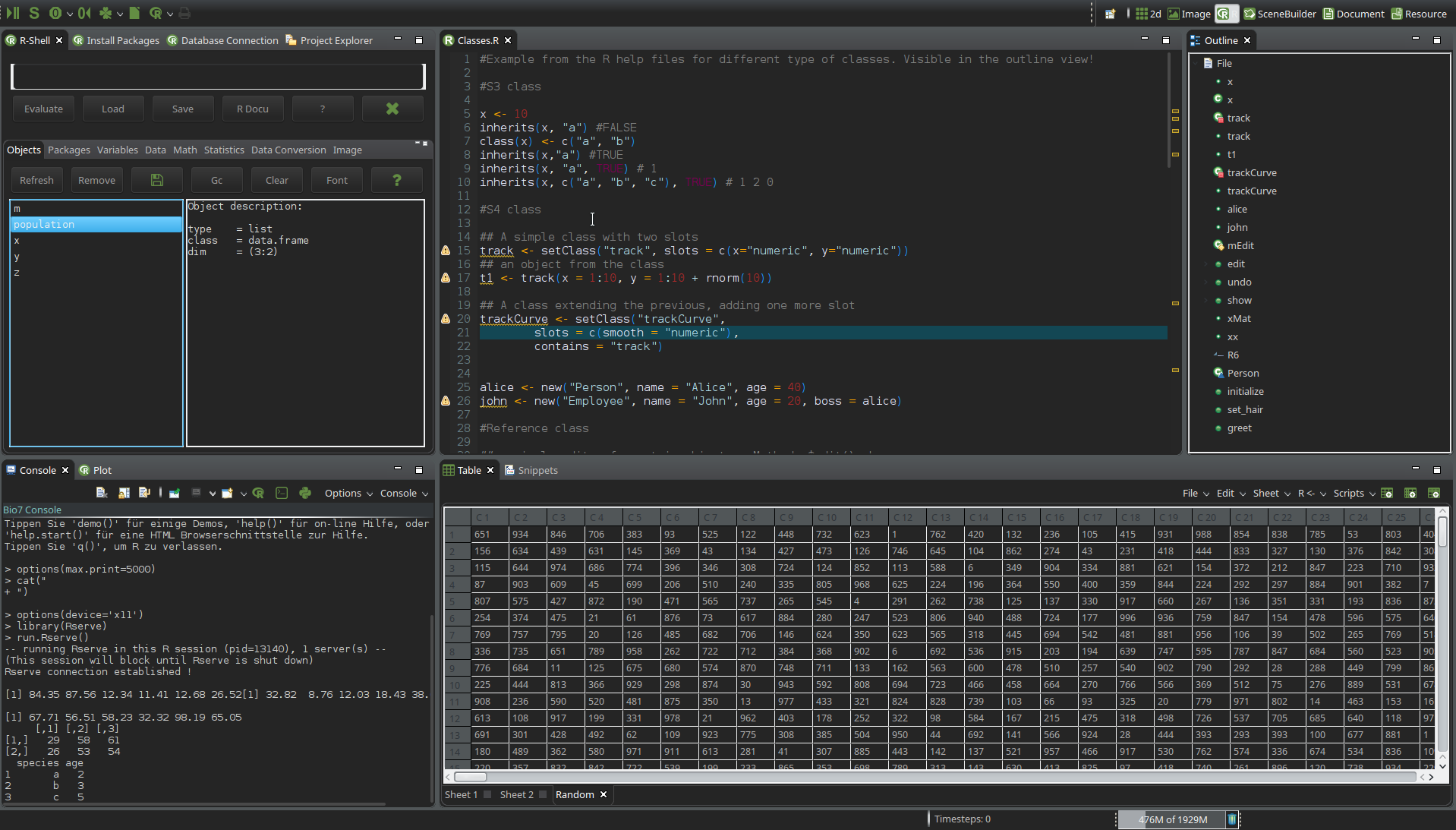
- Improved the default Bio7 theme (see Linux screenshot below)
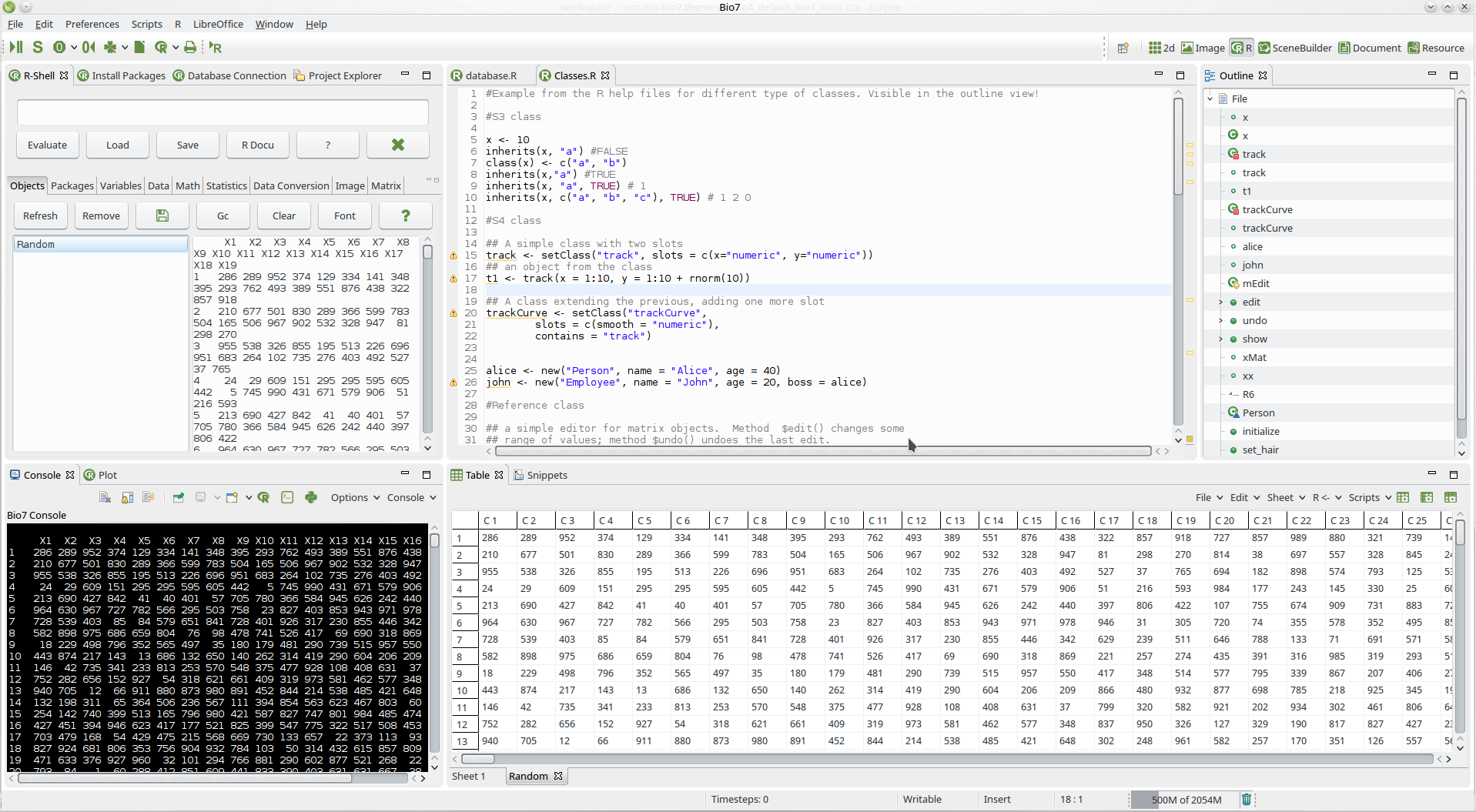
- Improved the theming of some Bio7 views (e.g., tables, text editors)
- Added a new shortcut for Windows and Linux to show or hide the Bio7 main menus – CTRL(CMD)+2, CTRL(CMD)+3). By default the application frame, toolbar and the status bar can be hidden, too. (see screenshot below with hidden window and menu – set the key shortcuts in the preferences!)
- Bio7 2.6 is based on Eclipse 4.7.1a
- Added the ‘Project Explorer’ view to many perspectives and deleted the ‘Navigator’ view in some of the perspectives (the ‘Project Explorer’ view displays errors and warnings and supports pasting of clipboard Java code, etc.)
- Added a break and interrupt action for the windows shell to end ,e.g., a ssh connection (CTRL+BREAK) or interrupt an ongoing shell process (CTRL+C)
- Added an API to send commands to the Windows/Linux/MacOSX shell
- Added SSH for Windows
- Added the embedded SSH application path to the shell
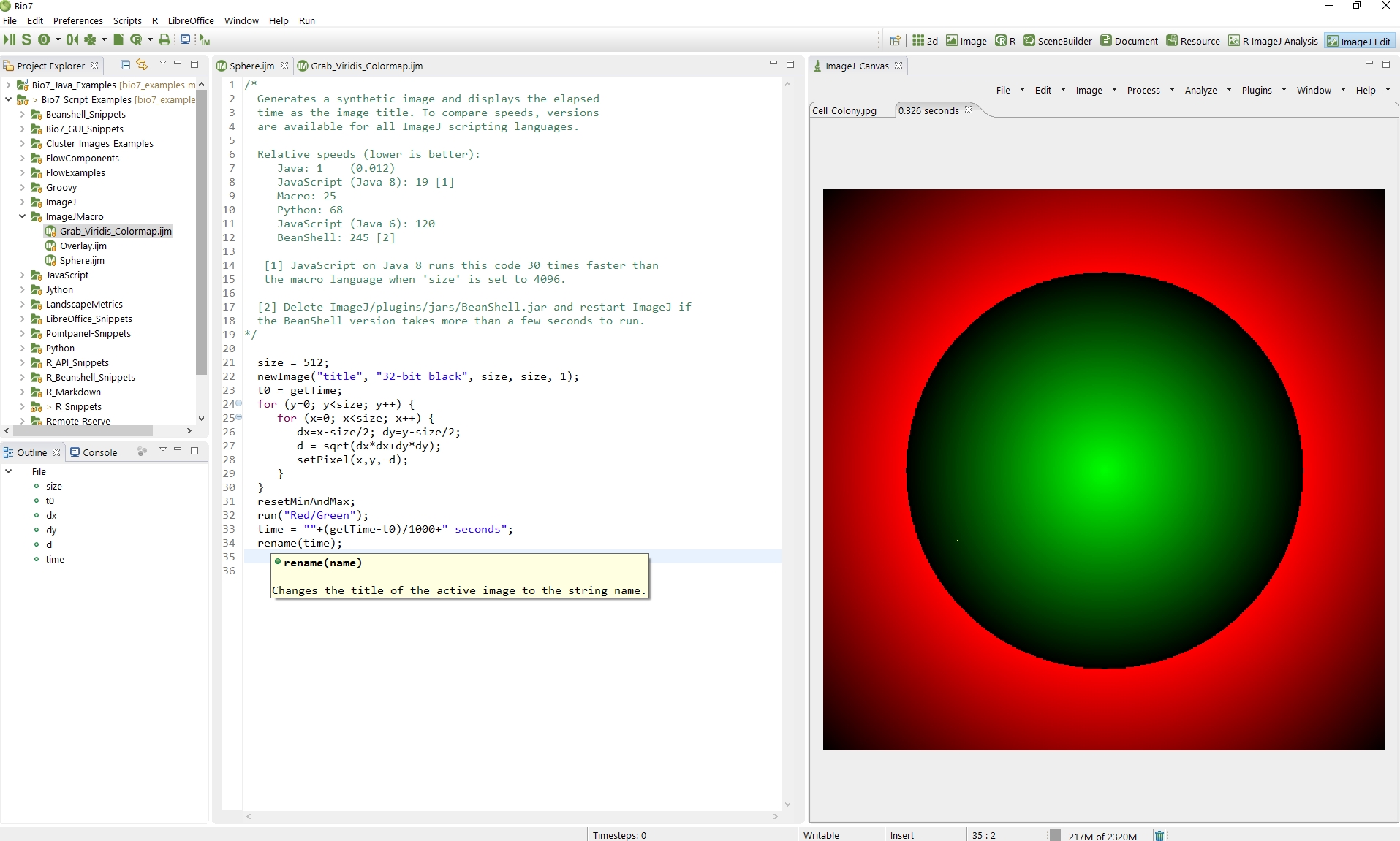
- Added a new ‘ImageJ Edit’ perspective to execute ImageJ scripts, macros side by side with the ImageJ-Canvas (see ImageJ section below)
- Added a easy to use R API for Java/BeanShell/Jython/Groovy/JavaScript to execute R commands or scripts in an extra job (thread)
- Improved several import/export scripts (now with Groovy) with the new Bio7 R API.
- Improved the Linux shell (added the default xterm path)
- The R package install script now works with a native R shell connection.
- Shiny applications can now be tested from all folders (not only project folder – e.g.; from Git folders)
R Editor
- Added an action to surround selected code with a “try and catch” skeleton
- Added an Project view/Navigator view popup action to start a shiny app in a arbitrary folder
- Added an action to open and insert a hex color in the editor
- Added a hoover dialog to display and replace selected hex and R colors (see screenshot below)
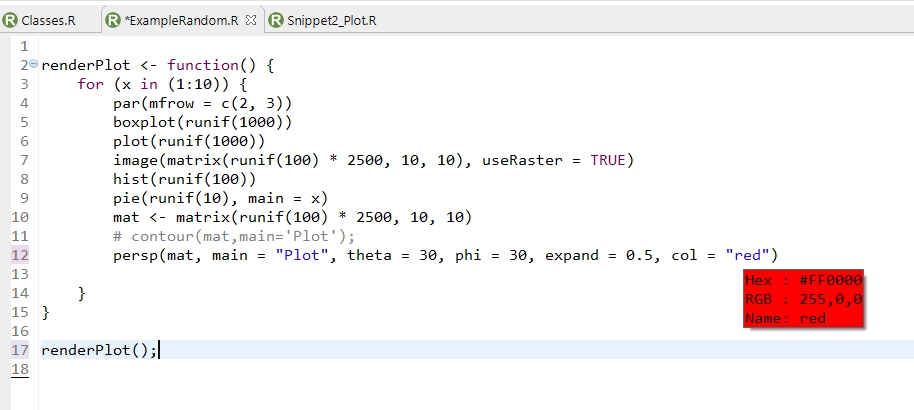
- Added an action in the hoover dialog to select hex colors
- Added an action in the hoover dialog to select default R colors by name (see screenshot below)
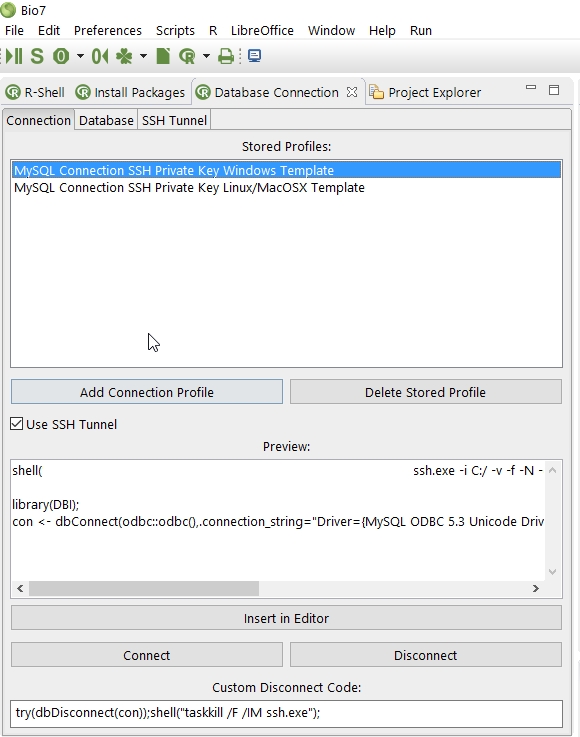
- Added a new database view to the R perspectives to store or apply database profiles and connections
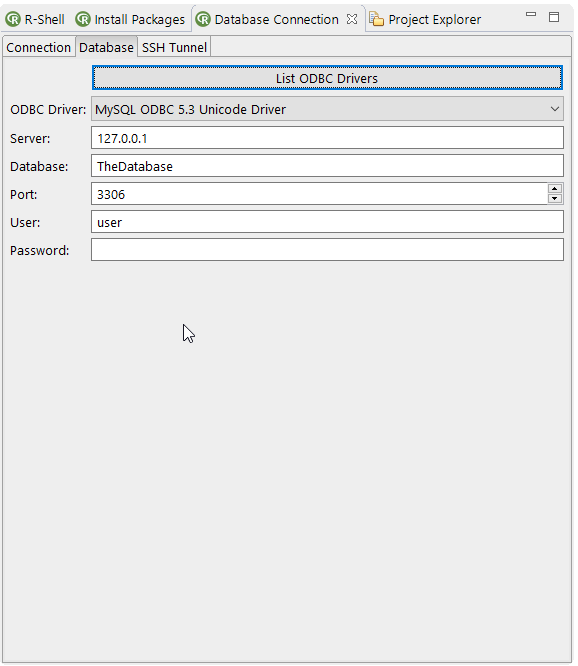
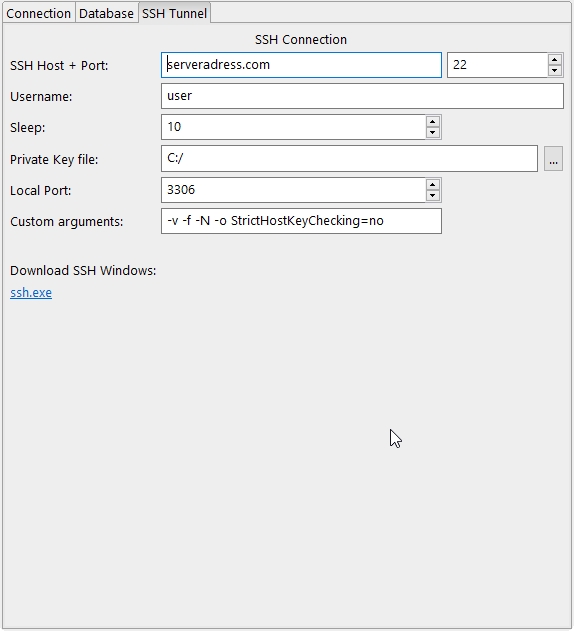
- Added SSH support for the database view (also for Windows!). Bio7 comes bundled with the latest SSH version of Windows
- Improved the code completion of the R-Shell
- Improved the default R package install script for Bio7
- Restructured the R editor context menu
ImageJ
- Added a full featured ImageJ macro editor with the following features:
- Dynamic error detection (grammar derived from the ECMA grammar by Bart Kiers)
- Toolbar action to execute ImageJ macros
- Syntax highlightening
- Editor debugging support (reusing and implementing interfaces and methods from – same keyboard shortcuts)
- Code completion (with browser help)
- Code templates (add you own templates)
- Info popups (text hoover)
- Outline view (var variables, variables, methods and macros)
- Automatic closing of braces, parentheses and strings
- Automatic indention of functions,loops, etc.
- Code folding
- Code formatting (based on the Eclipse-javascript-formatter by Sebastian Moran)
- Resize fonts functions (with Ctrl++ or CMD++, CTRL+- or CMD +-)
- Font preferences
- Mark occurences (scope independant)
- A copy function for the ImageJ forum
- The ImageJ view menu is now right aligned and foldable to the left
- Updated ImageJ to version 1.51r
- Several bugfixes for the ImageJ compatibility
Download and Installation:
Windows:
Just download the *.zip distribution file from http://bio7.org and unzip it in your preferred location. Bio7 comes bundled with a Java Runtime Environment, R and Rserve distribution and works out of the box.
Linux:
Download and extract the installation file from http://bio7.org.
For Linux you have to install R and Rserve.
To install Rserve open the R shell and then execute the menu action “Options->Install Rserve (coop. mode)”. This will download an install Rserve in your default R library location, see video below (please make sure that your default Linux R library install location has writing permissions!).
The special version of Rserve can also be downloaded here:
https://bitbucket.org/maustenfeld/bio7-new/downloads
For a manual installation in the R prompt type the following command to install the compiled package (replace with your file path!):
install.packages(“Users/yourName/Downloads/Rserve_1.8-4_Mac_cooperative.tgz”, repos=NULL)
Installation of Useful R Packages
R packages which are useful in combination with Bio7 can easily be installed with the main menu “Scripts” action:
Scripts->R-Scripts->InstallDefaultRPackages
Bio7 Documentation
For more information about Bio7 please consult the soon updated Bio7 User Guide.
A plethora of Bio7 videotutorials can be found on YouTube.
3 thoughts on “Bio7 2.6 for Windows and Linux Released”