07.08.2013
A new Windows version of Bio7 is available. This version comes with a lot of new features and improvements for Java, R and ImageJ. One highlight is that you can now interpret Jython (Python) code with Bio7. In addition a new console implementation is available which offers access to a native shell, different Java interpreted languages, etc.
Below you find new videos about some new features of Bio7 1.7, a general feature overview and installation details.
Videos:
Particle Analysis with ImageJ, R and the Points panel
Interpret Python scripts in Blender interactively
New features:
General
- Updated RCP platform to Eclipse 3.8.2
- Custom views can now be created with ID’s for Custom views inside of Bio7.
- Updated ImageJ
- Updated R
- Updated Rserve
- Changed the OpenOffice API and actions to LibreOffice
- New file wizards available (Jython, Html, Knitr, JavaFX, etc.)
- Removed the BeanShell menu (see section scripts)
- Removed the BeanShell console (now available as part of the Bio7 console)
- Extended the console of Bio7 (see section Scripts)
- Added Console options and typical commands
- Added console text options
- Integrated a new webeditor (JavaFX) which can be opened as a regular editor in Bio7
- Added new libraries for the support for e.g. serial communication (useful for Arduino boards)
- New examples available (serial examples, Jython, Multiple classes, etc.)
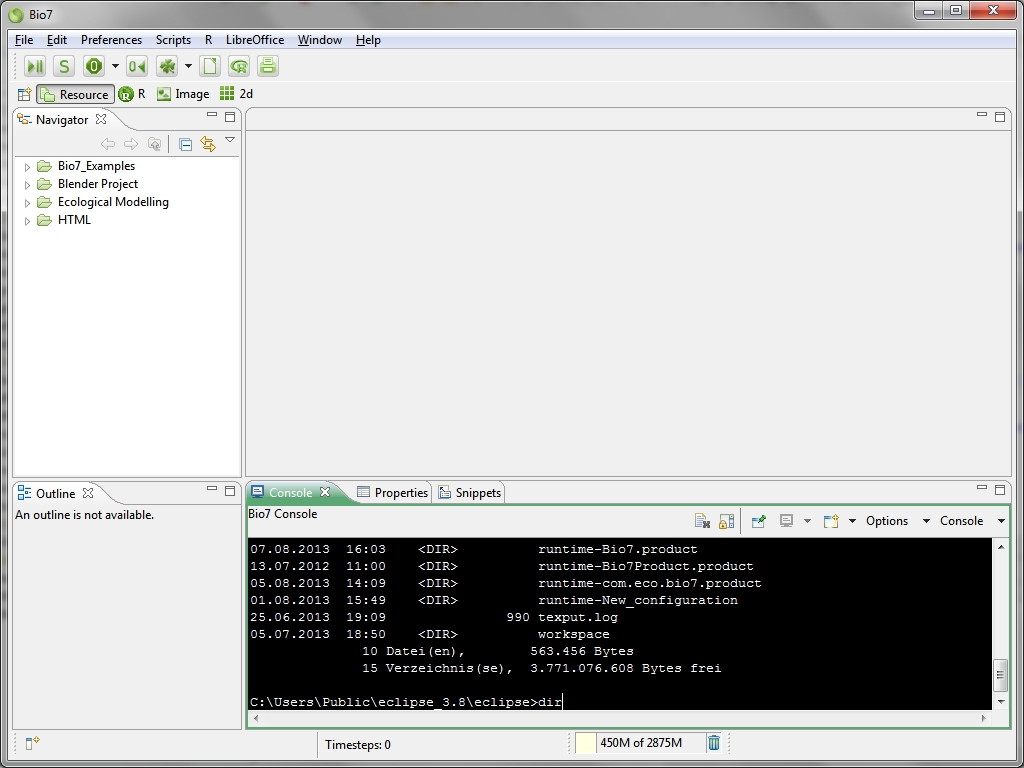
- Added native shell access to the Bio7 console (see image below)
- Added command templates to the shell menu
- Improved R console messages
- Updated embedded scientific Java libraries
- Now an Bio7 editor can add code snippets from selections to the Snippets view
- Rearranged the Image-Methods view
- Improved the Linux R output
- Startup scripts are now interpreted by Groovy for a custom startup of Bio7 (if enabled in the preferences)
- Added some new API methods (e.g. function to write to the new console).
- Rearranged and renamed some preferences for an improved overview.
- Added editor menus for the language specific preferences.
- Cleaned up the popup menu in the Navigator view and removed old BeanShell actions
- Native Python execution from the shell, Jython editor and the Flow editor can be enabled in the Bio7 preferences
- Native R execution from the shell, R editor, some R actions and Flow can be enabled in the Bio7 preferences
- Updated all native execution methods to use the recommended ProcessBuilder for Java >=1.7.21
- Resolved and fixed bugs
- Added preferences for Python, R and Blender to execute native R, Python, Blender Python scripts if enabled in the Bio7 preferences.
- Added default options for Python, Blender and R.
- Added new examples for Bio7 1.7
Java
- Dynamic compilation support for multiple Java classes and subclasses
- Compilation to *.class files is now supported for multiple Java classes
- If a *.class is selected and a main method is available it can be executed with a new action in the context menu of the Navigator view.
- Error indication support for multiple classes
- Drag and drop support for full classes stored along with grid definitions
- Added a new option to compile Java classes or classbody files in the Bio7 preferences
- JRE ext directory can be used to extend the Java classpath with external Java libs
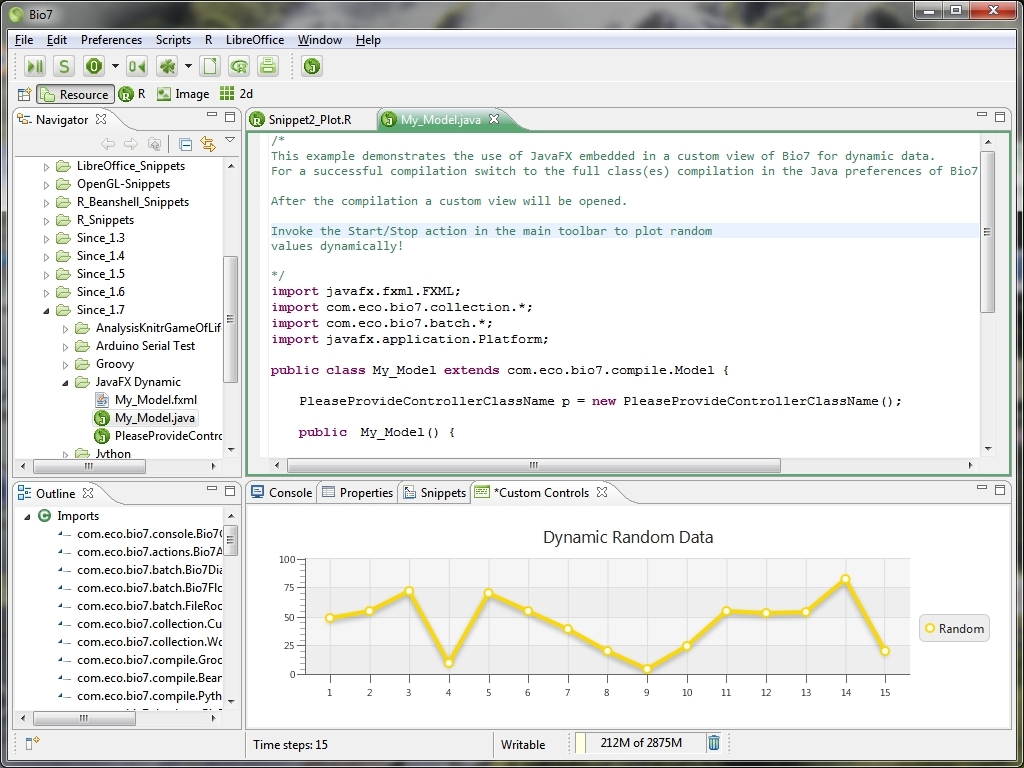
- JavaFX support for Bio7
- *.fxml files can now be created in the Bio7 workspace (along with a main Java file and a Controller file for the free available JavaFX Scene Builder from Oracle, see image below)
- Support for JavaFX Java source files
- JavaFX Scenes can be embedded into custom views
- Fullscreen support for JavaFX views (F1 = Fullscreen, F2= Fullscreen frame, F3=Fullscreen frame secondary monitor, F4= Fullscreen frame tertiary monitor, F5= Fullscreen frame quartiary monitor)
- If Fullscreen is activated an info message appears in the view which is in Fullscreen mode.
- Java files and subfiles can be instantiated and packages will be recognized, too (only if the file is opened in the Java editor and compiled with the editor action. Compilation from the context menu is available, too (without package recognition!)
- New Java wizards available (JavaFX, etc.)
- The main class of a Java project (which possible consists of multiple dependent subclasses) can be dragged in the Flow editor for execution.
- Added an action to compile multiple Java classes (context menu Navigator view)
- Added several compiler options to the Bio7 Java Preferences
- Added an editor context submenu to open the Java preferences faster
- The Constructor of a Java file is executed after compilation if available.
- If a main method of a regular Java class is available (not extending the Model class of Bio7!) it will be called after the compilation and instantiated.
Scripts
- Extended the default console to accept input for different Script interpreters
- BeanShell, Groovy and Jython commands can be evaluated in the new console
- Added a function to show a history of past commands (arrow keys)
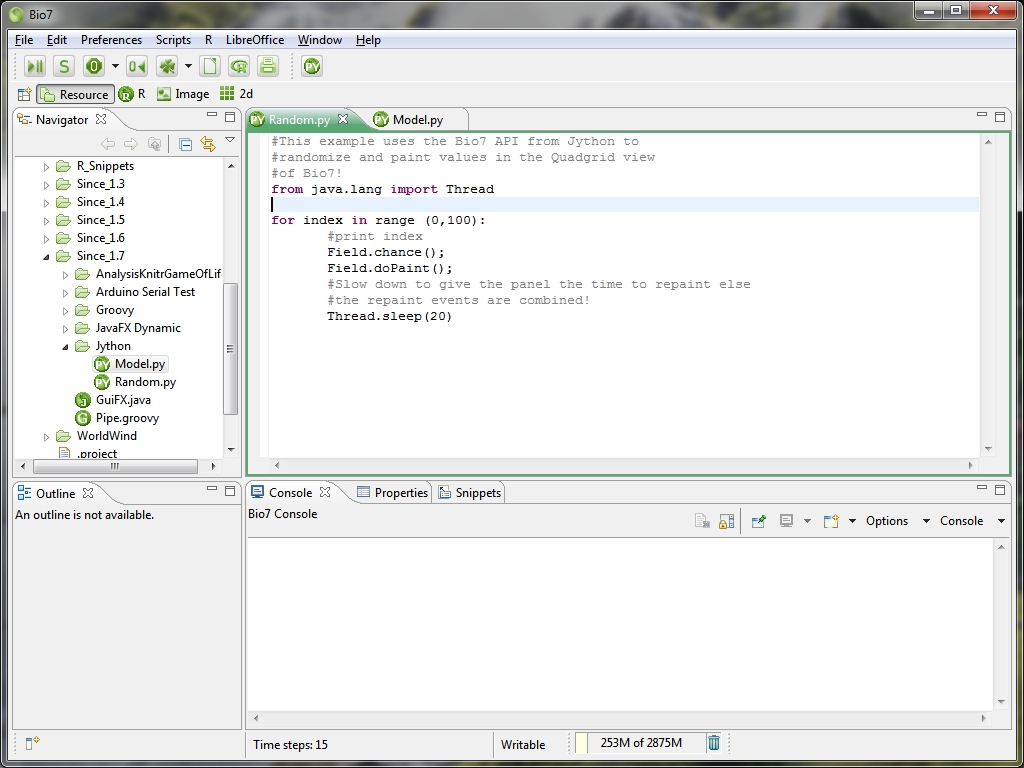
- Bio7 now supports the Jython interpreter
- New Jython editor available (see image below)
- Added new icons for the different editors
- Removed the old BeanShell console
- Removed the old BeanShell menu in the main menubar of Bio7
- Jython scripts can now be evaluated in the Flow editor
- Jython scripts can use the Bio7 API
- New file wizards for the different scripting languages available
- Python scripts can be executed if the native execution is enabled in the Bio7 preferences and the path to Python has been specified in the preferences
- Blender Python scripts can be executed if the native execution is enabled in the Bio7 preferences and the path to Blender has been specified in the preferences.
R
- Knitr support to compile knitr files (*.html, *.latex, etc.) in the Navigator of Bio7 (context menu)
- Knitr support in the Flow editor of Bio7 (for example to generate a report of simulation results, see image below)
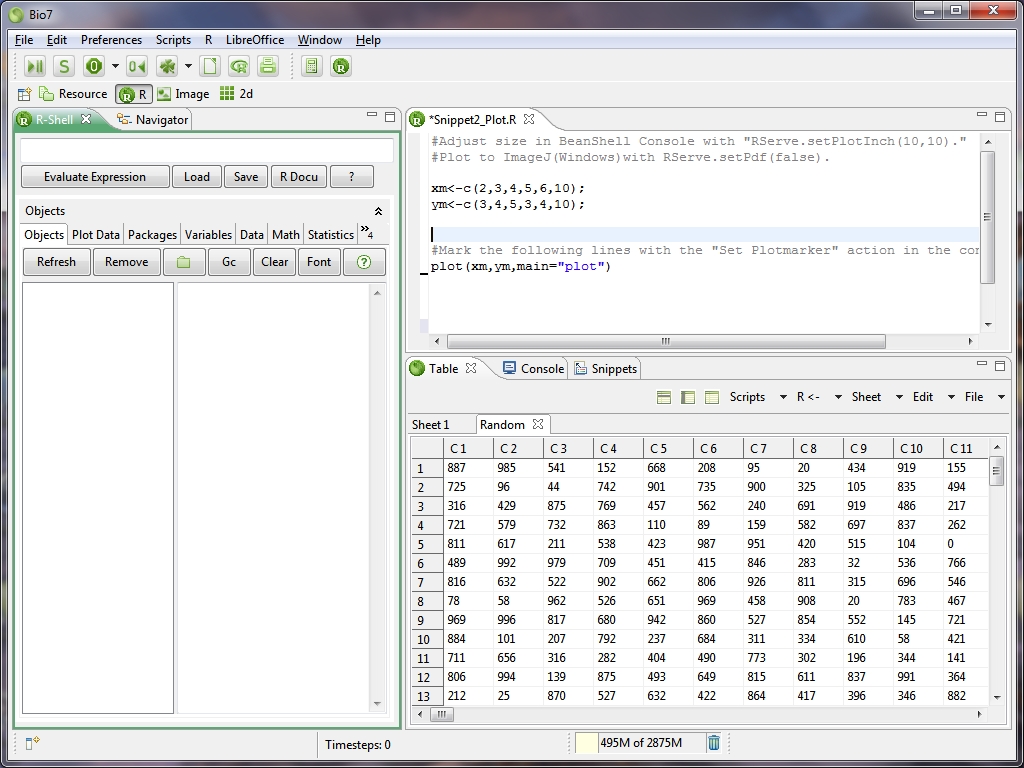
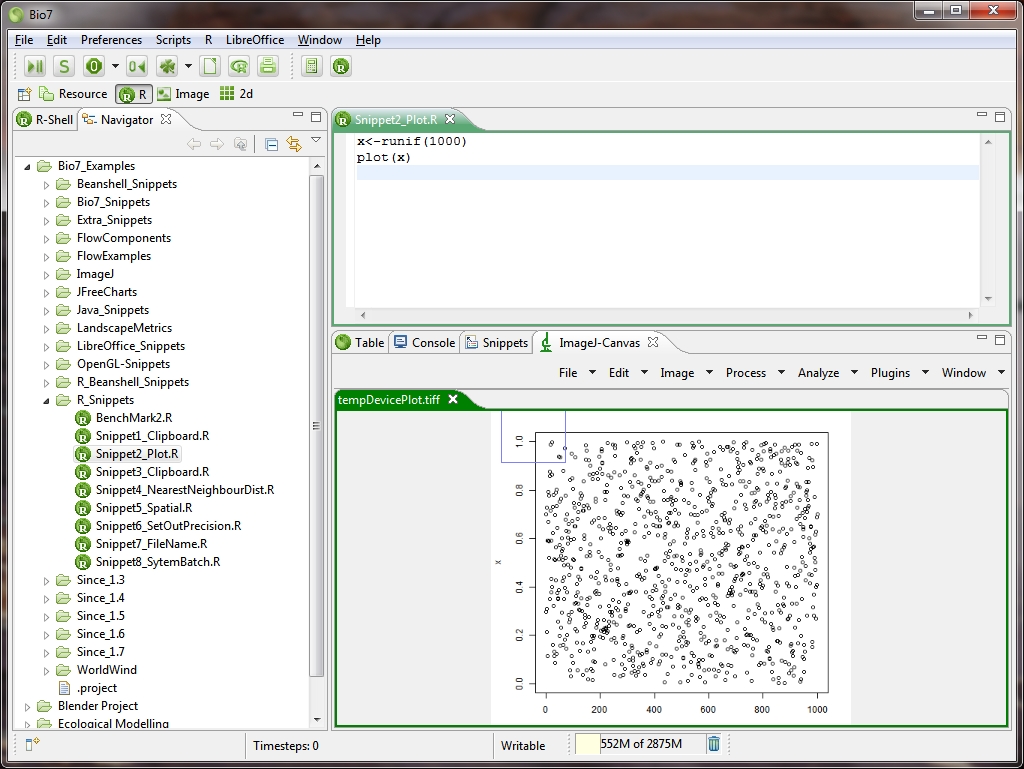
- Plots can be created interactively with the shell or the editor and are opened in ImageJ (as a plot display)
- New option in the Bio7 Rserve preferences to change the definition of the default plotting device.
- Added a new option to start Rserve automatically after Bio7 startup
- Experimental support to activate plots (Keys: Alt+a) in a blocking job as long as plots are opened (the R script for this functionality can be accessed and if necessary improved).
- Support for the interaction with commands (Windows) which require console commands (e.g. scan() – if the console requires input the textfield will be marked with a different color).
- Faster „Image“ plot in the plot section of the R-Shell view.
- Added a custom menu for scripts (R, Groovy, Jython, BeanShell) in the R-Shell view menu . Scripts which can be dropped in a special Bio7 folder
- Added a submenu in the context menu of the R editor to open the R preferences faster
- Added R package skeleton action in the context menu of the Navigator view to create R packages (with Rserve).
- Added R package built and install actions (with the native shell).
- Added R action to create *.pdf files out of *.Rd files (with the native shell)
- Added options in the Bio7 preferences for the built package commands
- Added native R access without the necessity to use Rserve. The editor, the shell and the Flow editor can be used with this new feature.
- Added fast workspace transfer function (F1-Key) from Rserve to the native R workspace
- Added Drag and Drop *.Rdata file feature for the native R access.
- Added Bio7 preferences options to enable the use of the native startup of R
- Added R and Rserve startup arguments in the Bio7 preferences.
- Added a submenu in the packages tab of the R-Shell view to refresh the view of available packages.
- Rearranged the R perspective and added the Editor Area, FileNavigator and Console view (ImageJ will be opened, too if a plot command occurs, see images below)
- If a plot command occurs the ImageJ-View will be opened with the plot as a default plotting device.
R – ImageJ
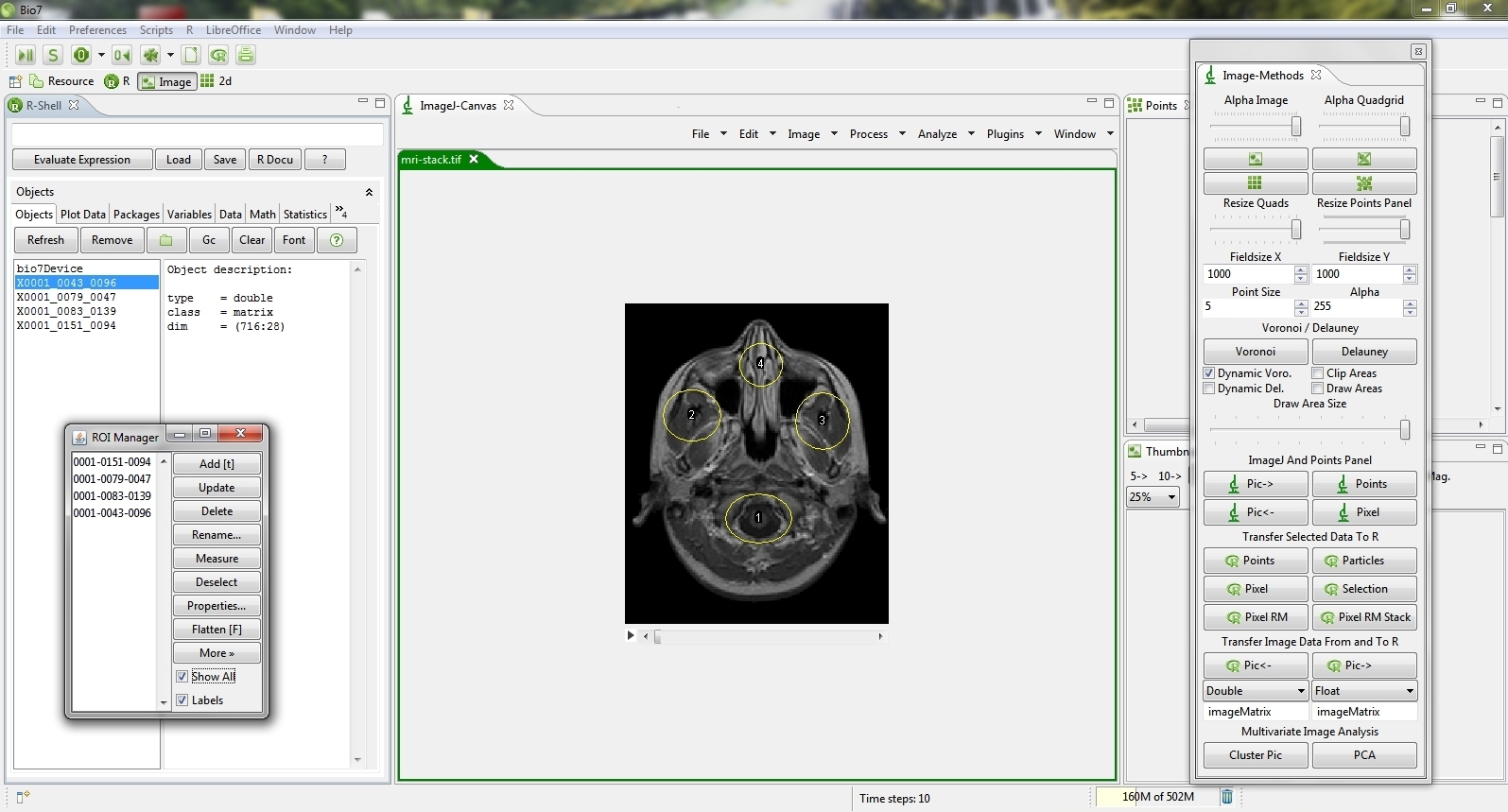
- Added a new method to transfer multiple selections from all or selected slices of a Stack (see image below)
- Splitted the pixel selection transfer action into two different actions for a better usability.
- Improved the Particles transfer method. Now uses the default ImageJ options.
Points view
- Points panel is now updated when Reset field action etc. is pressed.
Webeditor
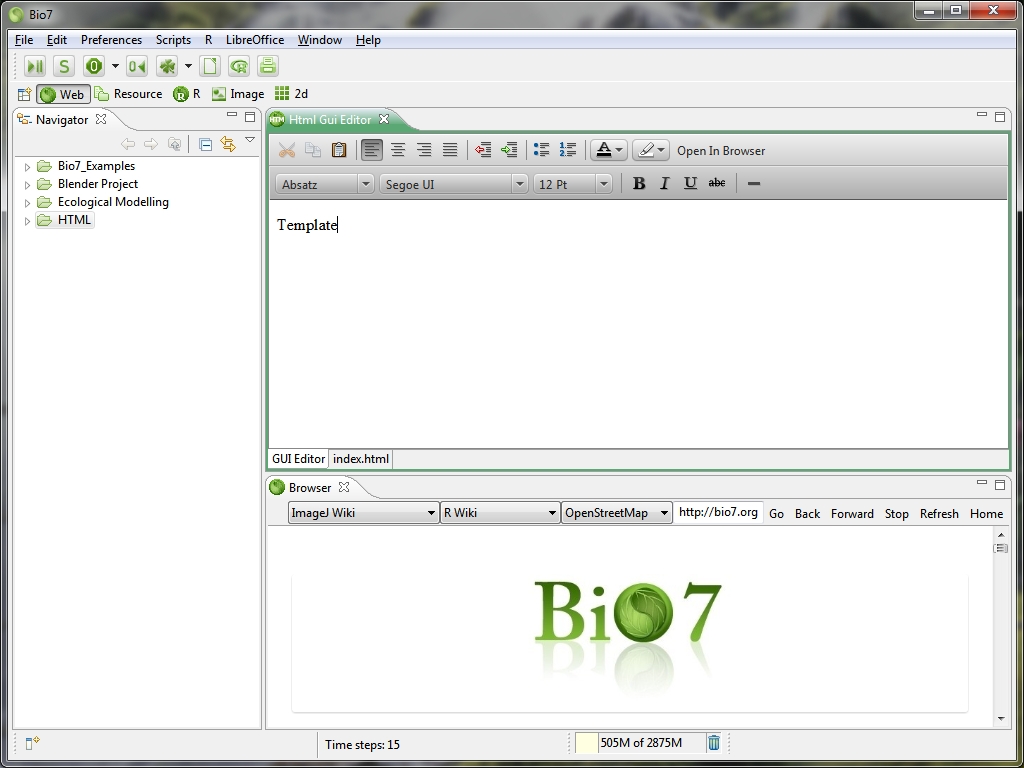
- Added a new web editor with source and GUI support (the GUI part uses the JavaFX web editor)
- Webeditor is now a regular Bio7 editor
- New file wizards available to create *.html files along with *javascript files
- Added action in the GUI to open the created file in the system web browser
Flow editor
- Gridsize for the layout can be configured
- Flow editor can now execute Jython scripts
- Flow editor can now execute either native python or Blender python scripts (if enabled in the preferences)
- Flow editor can now execute knitr *.Rhtml reports
- Flow editor can now execute knitr *.Rtex reports
- Flow editor can now execute a Java main class (dependant subclasses will automatically compiled, too)
WorldWind
- Improved the warnings for the access of the WorldWind API (if the WorldWind view is not active).
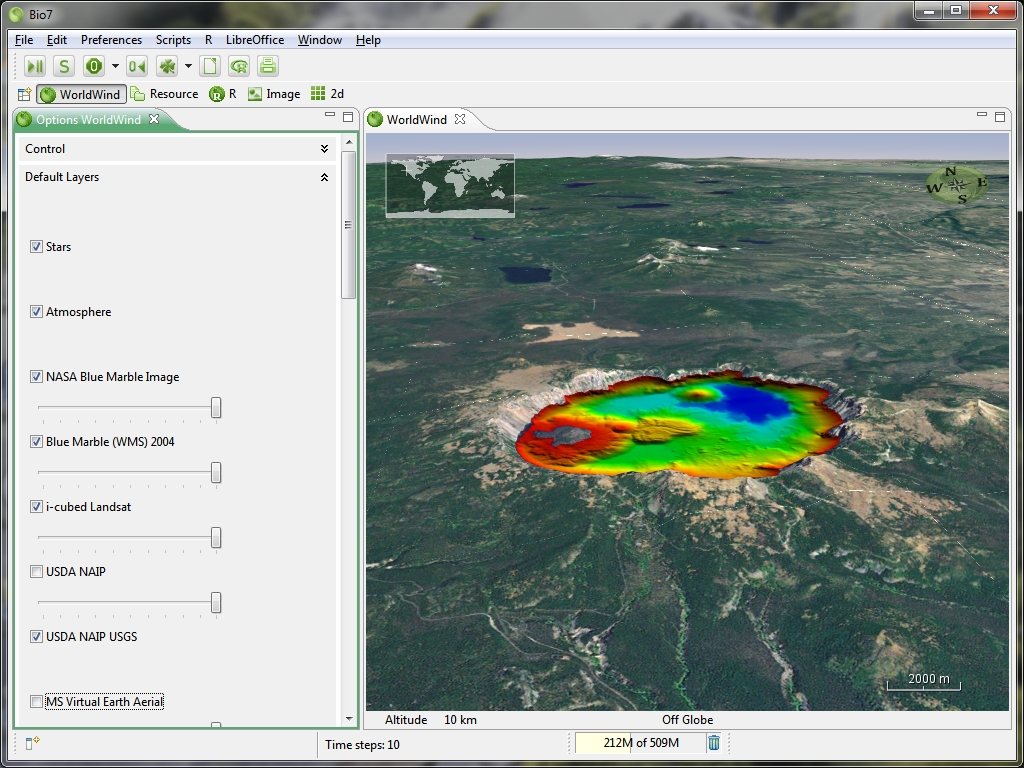
- Updated WorldWind to version 2.0 (using JOGL 2.0)
- Added a function to capture an animation to an ImageJ stack in a Bio7 job.
- Added options to adjust the framerate and the amount of frames to be rendered.
- Added WorldWind GeoTIFF import (coordinates will be set to the textfields)
- Added a „Go to” function for the GeoTIFF
- Added a first WorldWind Shapefile import
3d View
- Updated OpenGL library to JOGL 2.0
- Updated all Bio7 OpenGL examples to be compatible with JOGL 2.0
Bug Fixes
- Improved overall stability
- Fixed memory bugs with SWT
Installation Windows
The installation of Bio7 is similar to the installation of the Eclipse environment. Simply decompress the downloaded *.zip file in a preferred location on your file system. After decompressing with a standard zip-tool (like WinZip, Win Rar) the typical file-structure of an Eclipse based application will be created.
To start the application simply double click on the Bio7.exe (Windows) .
Optional R packages
In Bio7 1.7 please install the R package rgdal which is needed for some import and export scripts and knitr to use the report features with R.
For further information please read the Bio7 documentation on the Bio7 website.
Info about system requirements
It is recommended that your computer should have at least 2048 mb ram and a 1 ghz processor. A 3d Graphics Card which is OpenGL enabled (only necessary for the “Spatial” view and the embedded “WorldWind” view. To use the LibreOffice feature of Bio7 an installation of OpenOffice (LibreOffice) >= 2.0 is required. The path to LibreOffice can be adjusted in the Bio7 preferences to the default path.